Get Started in 10 Minutes#
In this page we will be showing basic capabilities of MDIO.
For demonstration purposes, we will download the Teapot Dome open-source dataset. The dataset details and licensing can be found at the SEG Wiki.
We are using the 3D seismic stack dataset named filt_mig.sgy.
The full link for the dataset (hosted on AWS): http://s3.amazonaws.com/teapot/filt_mig.sgy
Warning
For plotting, the notebook requires Matplotlib as a dependency. Please install it before executing using
pip install matplotlib or conda install matplotlib.
Downloading the SEG-Y Dataset#
Let’s download this dataset to our working directory. It may take from a few seconds up to a couple minutes based on your internet connection speed. The file is 386 MB in size.
The dataset is irregularly shaped, however it is padded to a rectangle with zero (dead traces). We will see that later at the live mask plotting.
from os import path
from urllib.request import urlretrieve
url = "http://s3.amazonaws.com/teapot/filt_mig.sgy"
urlretrieve(url, "filt_mig.sgy")
('filt_mig.sgy', <http.client.HTTPMessage at 0x7feb830b3b50>)
Ingesting to MDIO Format#
To do this, we can use the convenient SEG-Y to MDIO converter.
The inline and crossline values are located at bytes 181 and 185. Note that this is not SEG-Y standard.
from mdio import segy_to_mdio
segy_to_mdio(
segy_path="filt_mig.sgy",
mdio_path_or_buffer="filt_mig.mdio",
index_bytes=(181, 185),
index_names=("inline", "crossline"),
)
It only took a few seconds to ingest, since this is a very small file.
However, MDIO scales up to TB (that’s 1000 GB) sized volumes!
Opening the Ingested MDIO File#
Let’s open the MDIO file with the MDIOReader.
We will also turn on return_metadata function to get the live trace mask and trace headers.
from mdio import MDIOReader
mdio = MDIOReader("filt_mig.mdio", return_metadata=True)
Querying Metadata#
Now let’s look at the Textual Header by the convenient text_header attribute.
You will notice the text header is parsed as a list of strings that are 80 characters long.
mdio.text_header
['C 1 CLIENT: ROCKY MOUNTAIN OILFIELD TESTING CENTER ',
'C 2 PROJECT: NAVAL PETROLEUM RESERVE #3 (TEAPOT DOME); NATRONA COUNTY, WYOMING ',
'C 3 LINE: 3D ',
'C 4 ',
'C 5 THIS IS THE FILTERED POST STACK MIGRATION ',
'C 6 ',
'C 7 INLINE 1, XLINE 1: X COORDINATE: 788937 Y COORDINATE: 938845 ',
'C 8 INLINE 1, XLINE 188: X COORDINATE: 809501 Y COORDINATE: 939333 ',
'C 9 INLINE 188, XLINE 1: X COORDINATE: 788039 Y COORDINATE: 976674 ',
'C10 INLINE NUMBER: MIN: 1 MAX: 345 TOTAL: 345 ',
'C11 CROSSLINE NUMBER: MIN: 1 MAX: 188 TOTAL: 188 ',
"C12 TOTAL NUMBER OF CDPS: 64860 BIN DIMENSION: 110' X 110' ",
'C13 ',
'C14 ',
'C15 ',
'C16 ',
'C17 ',
'C18 ',
'C19 GENERAL SEGY INFORMATION ',
'C20 RECORD LENGHT (MS): 3000 ',
'C21 SAMPLE RATE (MS): 2.0 ',
'C22 DATA FORMAT: 4 BYTE IBM FLOATING POINT ',
'C23 BYTES 13- 16: CROSSLINE NUMBER (TRACE) ',
'C24 BYTES 17- 20: INLINE NUMBER (LINE) ',
'C25 BYTES 81- 84: CDP_X COORD ',
'C26 BYTES 85- 88: CDP_Y COORD ',
'C27 BYTES 181-184: INLINE NUMBER (LINE) ',
'C28 BYTES 185-188: CROSSLINE NUMBER (TRACE) ',
'C29 BYTES 189-192: CDP_X COORD ',
'C30 BYTES 193-196: CDP_Y COORD ',
'C31 ',
'C32 ',
'C33 ',
'C34 ',
'C35 ',
'C36 Processed by: Excel Geophysical Services, Inc. ',
'C37 8301 East Prentice Ave. Ste. 402 ',
'C38 Englewood, Colorado 80111 ',
'C39 (voice) 303.694.9629 (fax) 303.771.1646 ',
'C40 END EBCDIC ']
MDIO parses the binary header into a Python dictionary.
We can easily query it by the binary_header attribute and see critical information about the original file.
Since we use segyio for parsing the SEG-Y, the field names conform to it.
mdio.binary_header
{'AmplitudeRecovery': 4,
'AuxTraces': 0,
'BinaryGainRecovery': 1,
'CorrelatedTraces': 2,
'EnsembleFold': 57,
'ExtAuxTraces': 0,
'ExtEnsembleFold': 0,
'ExtSamples': 0,
'ExtSamplesOriginal': 0,
'ExtendedHeaders': 0,
'Format': 1,
'ImpulseSignalPolarity': 1,
'Interval': 2000,
'IntervalOriginal': 0,
'JobID': 9999,
'LineNumber': 9999,
'MeasurementSystem': 2,
'ReelNumber': 1,
'SEGYRevision': 0,
'SEGYRevisionMinor': 0,
'Samples': 1501,
'SamplesOriginal': 1501,
'SortingCode': 4,
'Sweep': 0,
'SweepChannel': 0,
'SweepFrequencyEnd': 0,
'SweepFrequencyStart': 0,
'SweepLength': 0,
'SweepTaperEnd': 0,
'SweepTaperStart': 0,
'Taper': 0,
'TraceFlag': 0,
'Traces': 188,
'VerticalSum': 1,
'VibratoryPolarity': 0}
Fetching Data and Plotting#
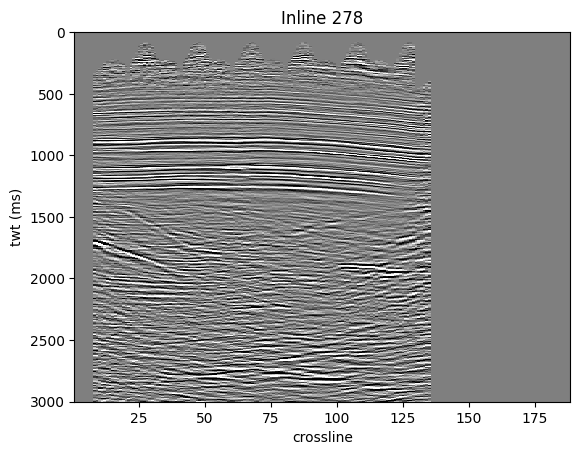
Now we will demonstrate getting an inline from MDIO.
Because MDIO can hold various dimensionality of data, we have to first query the inline location.
Then we can use the queried index to get the data itself.
We will also plot the inline, for this we need the crossline and sample coordinates.
MDIO stores dataset statistics. We can use the standard deviation (std) value of the dataset to adjust the gain.
import matplotlib.pyplot as plt
crosslines = mdio.grid.select_dim("crossline").coords
times = mdio.grid.select_dim("sample").coords
std = mdio.stats["std"]
inline_index = int(mdio.coord_to_index(278, dimensions="inline"))
il_mask, il_headers, il_data = mdio[inline_index]
vmin, vmax = -2 * std, 2 * std
plt.pcolormesh(crosslines, times, il_data.T, vmin=vmin, vmax=vmax, cmap="gray_r")
plt.gca().invert_yaxis()
plt.title(f"Inline {278}")
plt.xlabel("crossline")
plt.ylabel("twt (ms)");
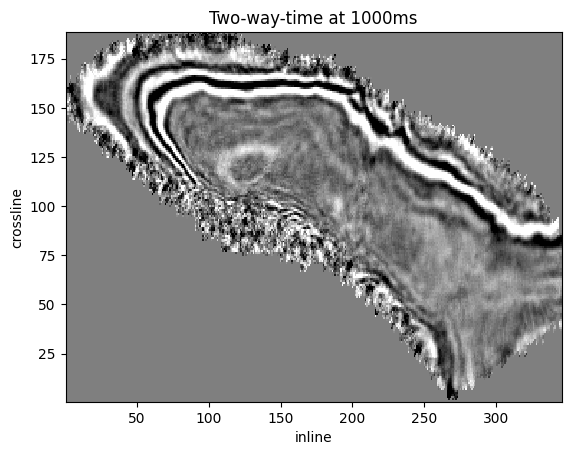
Let’s do the same with a time sample.
We already have crossline labels and standard deviation, so we don’t have to fetch it again.
We will display two-way-time at 1,000 ms.
inlines = mdio.grid.select_dim("inline").coords
twt_index = int(mdio.coord_to_index(1_000, dimensions="sample"))
z_mask, z_headers, z_data = mdio[:, :, twt_index]
vmin, vmax = -2 * std, 2 * std
plt.pcolormesh(inlines, crosslines, z_data.T, vmin=vmin, vmax=vmax, cmap="gray_r")
plt.title(f"Two-way-time at {1000}ms")
plt.xlabel("inline")
plt.ylabel("crossline");
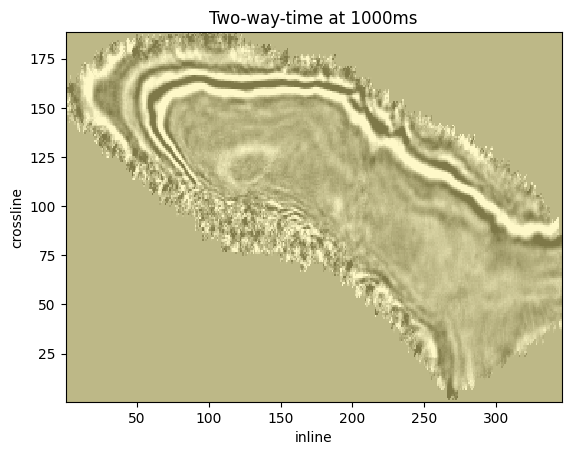
We can also overlay live mask with the time slice. However, in this example dataset is zero padded.
The live mask will always show True.
live_mask = mdio.live_mask[:]
plt.pcolormesh(inlines, crosslines, live_mask.T, vmin=0, vmax=1, alpha=0.5)
plt.pcolormesh(inlines, crosslines, z_data.T, vmin=vmin, vmax=vmax, cmap="gray_r", alpha=0.5)
plt.title(f"Two-way-time at {1000}ms")
plt.xlabel("inline")
plt.ylabel("crossline");
Query Headers#
We can query headers for the whole dataset very quickly because they are separated from the seismic wavefield.
Let’s get all the headers for byte 189 and 193 (X and Y in this dataset, non-standard).
Note that the header maps will still honor the geometry of the dataset!
mdio._headers[:]["189"]
array([[788937, 789047, 789157, ..., 809282, 809392, 809502],
[788935, 789045, 789155, ..., 809279, 809389, 809499],
[788932, 789042, 789152, ..., 809276, 809386, 809496],
...,
[788044, 788154, 788264, ..., 808389, 808499, 808609],
[788042, 788152, 788262, ..., 808386, 808496, 808606],
[788039, 788149, 788259, ..., 808383, 808493, 808603]], dtype=int32)
mdio._headers[:]["193"]
array([[938846, 938848, 938851, ..., 939329, 939331, 939334],
[938956, 938958, 938961, ..., 939439, 939441, 939444],
[939066, 939068, 939071, ..., 939549, 939551, 939554],
...,
[976455, 976458, 976460, ..., 976938, 976941, 976943],
[976565, 976568, 976570, ..., 977048, 977051, 977053],
[976675, 976678, 976680, ..., 977158, 977161, 977163]], dtype=int32)
or both at he same time:
mdio._headers[:][["189", "193"]]
array([[(788937, 938846), (789047, 938848), (789157, 938851), ...,
(809282, 939329), (809392, 939331), (809502, 939334)],
[(788935, 938956), (789045, 938958), (789155, 938961), ...,
(809279, 939439), (809389, 939441), (809499, 939444)],
[(788932, 939066), (789042, 939068), (789152, 939071), ...,
(809276, 939549), (809386, 939551), (809496, 939554)],
...,
[(788044, 976455), (788154, 976458), (788264, 976460), ...,
(808389, 976938), (808499, 976941), (808609, 976943)],
[(788042, 976565), (788152, 976568), (788262, 976570), ...,
(808386, 977048), (808496, 977051), (808606, 977053)],
[(788039, 976675), (788149, 976678), (788259, 976680), ...,
(808383, 977158), (808493, 977161), (808603, 977163)]],
dtype={'names': ['189', '193'], 'formats': ['<i4', '<i4'], 'offsets': [188, 192], 'itemsize': 232})
As we mentioned before, we can also get specific slices of headers while fetching a slice.
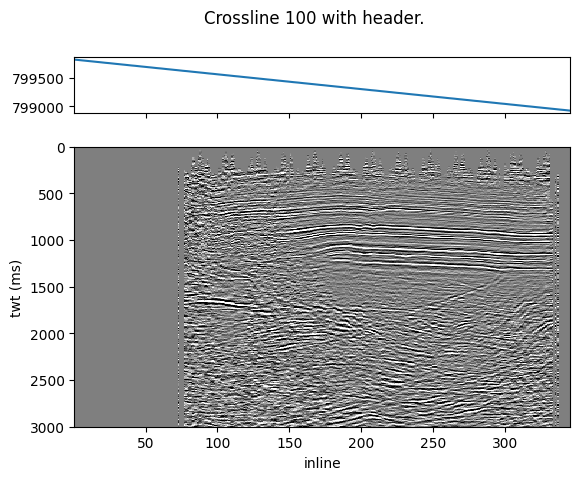
Let’s fetch a crossline, we are still using some previous parameters.
Since crossline is our second dimension, we can put the index in the second mdio[...] axis.
Since MDIO returns the headers as well, we can plot the headers on top of the image.
All headers will be returned, so we can select the X-coordinate at byte 189.
Full headers can be mapped and plotted as well, but we won’t demonstrate that here.
crossline_index = int(mdio.coord_to_index(100, dimensions="crossline"))
xl_mask, xl_headers, xl_data = mdio[:, crossline_index]
vmin, vmax = -2 * std, 2 * std
gs_kw = dict(height_ratios=(1, 5))
fig, ax = plt.subplots(2, 1, gridspec_kw=gs_kw, sharex="all")
ax[0].plot(inlines, xl_headers["189"])
ax[1].pcolormesh(inlines, times, xl_data.T, vmin=vmin, vmax=vmax, cmap="gray_r")
ax[1].invert_yaxis()
ax[1].set_xlabel("inline")
ax[1].set_ylabel("twt (ms)")
plt.suptitle(f"Crossline {100} with header.");
MDIO to SEG-Y Conversion#
Finally, let’s demonstrate going back to SEG-Y.
We will use the convenient mdio_to_segy function and write it out as a round-trip file.
from mdio import mdio_to_segy
mdio_to_segy(
mdio_path_or_buffer="filt_mig.mdio",
output_segy_path="filt_mig_roundtrip.sgy",
)
Array shape is (345, 188, 1501)
Setting (dask) chunks from (128, 128, 128) to (128, 128, 1501)
Validate Round-Trip SEG-Y File#
We can validate if the round-trip SEG-Y file is matching the original using segyio.
Step by step:
Open original file
Open round-trip file
Compare text headers
Compare binary headers
Compare 100 random headers and traces
import numpy as np
import segyio
original_fp = segyio.open("filt_mig.sgy", iline=181, xline=185)
roundtrip_fp = segyio.open("filt_mig_roundtrip.sgy", iline=181, xline=185)
# Compare text header
assert original_fp.text[0] == roundtrip_fp.text[0]
# Compare bin header
assert original_fp.bin == roundtrip_fp.bin
# Compare 100 random trace headers and traces
rng = np.random.default_rng()
rand_indices = rng.integers(low=0, high=original_fp.tracecount, size=100)
for idx in rand_indices:
np.testing.assert_equal(original_fp.header[idx], roundtrip_fp.header[idx])
np.testing.assert_equal(original_fp.trace[idx], roundtrip_fp.trace[idx])
original_fp.close()
roundtrip_fp.close()